Introduction
Overview
Teaching: 15 min
Exercises: 15 minQuestions
How can I make my results easier to reproduce?
Objectives
Understand our example problem.
Possible current method
Let’s imagine that we’re interested in seeing the frequency of various words in various books.
Files
Files for this section are available in the Github repository or available by downloading the zip or tar file, e.g.:
$ curl -O https://raw.githubusercontent.com/ARCCA/intro_nextflow/gh-pages/files/nextflow-lesson.zip
We’ve compiled our raw data i.e. the books we want to analyse and have prepared several Python scripts that together make up our analysis pipeline.
Let’s take quick look at one of the books using the command head books/isles.txt.
head books/isles.txt
By default, head displays the first 10 lines of the specified file.
A JOURNEY TO THE WESTERN ISLANDS OF SCOTLAND
INCH KEITH
I had desired to visit the Hebrides, or Western Islands of Scotland, so
long, that I scarcely remember how the wish was originally excited; and
was in the Autumn of the year 1773 induced to undertake the journey, by
finding in Mr. Boswell a companion, whose acuteness would help my
Our directory has the Python scripts and data files we we will be working with:
|- books
| |- abyss.txt
| |- isles.txt
| |- last.txt
| |- LICENSE_TEXTS.md
| |- sierra.txt
|- plotcount.py
|- wordcount.py
|- zipf_test.py
The first step is to count the frequency of each word in a book.
The first argument (books/isles.txt) to wordcount.py is the file to analyse,
and the last argument (isles.dat) specifies the output file to write.
$ python wordcount.py books/isles.txt isles.dat
Let’s take a quick peek at the result.
$ head -5 isles.dat
This shows us the top 5 lines in the output file:
the 3822 6.7371760973
of 2460 4.33632998414
and 1723 3.03719372466
to 1479 2.60708619778
a 1308 2.30565838181
We can see that the file consists of one row per word. Each row shows the word itself, the number of occurrences of that word, and the number of occurrences as a percentage of the total number of words in the text file.
We can do the same thing for a different book:
$ python wordcount.py books/abyss.txt abyss.dat
$ head -5 abyss.dat
the 4044 6.35449402891
and 2807 4.41074795726
of 1907 2.99654305468
a 1594 2.50471401634
to 1515 2.38057825267
Let’s visualise the results.
The script plotcount.py reads in a data file and plots the 10 most
frequently occurring words as a text-based bar plot:
$ python plotcount.py isles.dat ascii
the ########################################################################
of ##############################################
and ################################
to ############################
a #########################
in ###################
is #################
that ############
by ###########
it ###########
plotcount.py can also show the plot graphically:
$ python plotcount.py isles.dat show
Close the window to exit the plot.
plotcount.py can also create the plot as an image file (e.g. a PNG file):
$ python plotcount.py isles.dat isles.png
Finally, let’s test Zipf’s law for these books:
$ python zipf_test.py abyss.dat isles.dat
Book First Second Ratio
abyss 4044 2807 1.44
isles 3822 2460 1.55
Zipf’s Law
Zipf’s Law is an empirical law formulated using mathematical statistics that refers to the fact that many types of data studied in the physical and social sciences can be approximated with a Zipfian distribution, one of a family of related discrete power law probability distributions.
Zipf’s law was originally formulated in terms of quantitative linguistics, stating that given some corpus of natural language utterances, the frequency of any word is inversely proportional to its rank in the frequency table. For example, in the Brown Corpus of American English text, the word the is the most frequently occurring word, and by itself accounts for nearly 7% of all word occurrences (69,971 out of slightly over 1 million). True to Zipf’s Law, the second-place word of accounts for slightly over 3.5% of words (36,411 occurrences), followed by and (28,852). Only 135 vocabulary items are needed to account for half the Brown Corpus.
Source: Wikipedia:
Together these scripts implement a common workflow:
- Read a data file.
- Perform an analysis on this data file.
- Write the analysis results to a new file.
- Plot a graph of the analysis results.
- Save the graph as an image, so we can put it in a paper.
- Make a summary table of the analyses
Running wordcount.py and plotcount.py at the shell prompt, as we
have been doing, is fine for one or two files. If, however, we had 5
or 10 or 20 text files,
or if the number of steps in the pipeline were to expand, this could turn into
a lot of work.
Plus, no one wants to sit and wait for a command to finish, even just for 30
seconds.
The most common solution to the tedium of data processing is to write a shell script that runs the whole pipeline from start to finish.
Using your text editor of choice (e.g. nano), add the following to a new file named
run_pipeline.sh.
# USAGE: bash run_pipeline.sh
# to produce plots for isles and abyss
# and the summary table for the Zipf's law tests
python wordcount.py books/isles.txt isles.dat
python wordcount.py books/abyss.txt abyss.dat
python plotcount.py isles.dat isles.png
python plotcount.py abyss.dat abyss.png
# Generate summary table
python zipf_test.py abyss.dat isles.dat > results.txt
Run the script and check that the output is the same as before:
$ bash run_pipeline.sh
$ cat results.txt
This shell script solves several problems in computational reproducibility:
- It explicitly documents our pipeline, making communication with colleagues (and our future selves) more efficient.
- It allows us to type a single command,
bash run_pipeline.sh, to reproduce the full analysis. - It prevents us from repeating typos or mistakes. You might not get it right the first time, but once you fix something it’ll stay fixed.
Despite these benefits it has a few shortcomings.
Let’s adjust the width of the bars in our plot produced by plotcount.py.
Edit plotcount.py so that the bars are 0.8 units wide instead of 1 unit.
(Hint: replace width = 1.0 with width = 0.8 in the definition of
plot_word_counts.)
Now we want to recreate our figures.
We could just bash run_pipeline.sh again.
That would work, but it could also be a big pain if counting words takes
more than a few seconds.
The word counting routine hasn’t changed; we shouldn’t need to recreate
those files.
Alternatively, we could manually rerun the plotting for each word-count file. (Experienced shell scripters can make this easier on themselves using a for-loop.)
$ for book in abyss isles; do python plotcount.py $book.dat $book.png; done
With this approach, however, we don’t get many of the benefits of having a shell script in the first place.
Another popular option is to comment out a subset of the lines in
run_pipeline.sh:
# USAGE: bash run_pipeline.sh
# to produce plots for isles and abyss
# and the summary table
# These lines are commented out because they don't need to be rerun.
#python wordcount.py books/isles.txt isles.dat
#python wordcount.py books/abyss.txt abyss.dat
python plotcount.py isles.dat isles.png
python plotcount.py abyss.dat abyss.png
# This line is also commented out because it doesn't need to be rerun.
# python zipf_test.py abyss.dat isles.dat > results.txt
Then, we would run our modified shell script using bash run_pipeline.sh.
But commenting out these lines, and subsequently un-commenting them, can be a hassle and source of errors in complicated pipelines. What happens if we have hundreds of input files? No one wants to enter the same command a hundred times, and then edit the result.
What we really want is an executable description of our pipeline that allows software to do the tricky part for us: figuring out what tasks need to be run where and when, then perform those tasks for us.
What is Nextflow and why are we using it?
There are many different tools that researchers use to automate this type of work.
Nextflow has been chosen for this lesson:
-
It’s free, open-source, and installs in about 5 seconds flat via a script.
-
Fast prototyping by reusing existing scripts.
-
Reproducibility with technologies such as Docker and Singularity to reproduce other user’s work.
-
Portable due to being able to execute on different platforms such as common job schedulers and cloud platforms.
-
Unified parallelism based on dataflow programming model, expressing jobs by their inputs and outputs.
-
Continuous checkpoints to allow resuming and monitoring intermediate tasks.
-
Stream oriented by extending the standard Unix pipes model with its fluent DSL.
Snakemake is another very popular tool, and similar lesson can be found in Carpentry HPC Python, for several reasons:
-
It’s free, open-source, and installs in about 5 seconds flat via
pip. -
Snakemake works cross-platform (Windows, MacOS, Linux) and is compatible with all HPC schedulers. More importantly, the same workflow will work and scale appropriately regardless of whether it’s on a laptop or cluster without modification.
-
Snakemake uses pure Python syntax. There is no tool specific-language to learn like in GNU Make, NextFlow, WDL, etc.. Even if students end up not liking Snakemake, you’ve still taught them how to program in Python at the end of the day.
-
Anything that you can do in Python, you can do with Snakemake (since you can pretty much execute arbitrary Python code anywhere).
-
Snakemake was written to be as similar to GNU Make as possible. Users already familiar with Make will find Snakemake quite easy to use.
-
It’s easy. You can (hopefully!) learn Snakemake in an afternoon!
The rest of these lessons aim to teach you how to use Nextflow by example. Our goal is to automate our example workflow, and have it do everything for us in parallel regardless of where and how it is run (and have it be reproducible!).
Key Points
Bash scripts are not an efficient way of storing a workflow.
Nextflow is one method of managing a complex computational workflow.
The basics
Overview
Teaching: 15 min
Exercises: 15 minQuestions
How do I write a simple workflow?
Objectives
Understand the files that control Nextflow.
Understand the components of Nextflow: processes, channels, executors, and operators.
Write a simple Nextflow script.
Run Nextflow from the shell.
Software modules
Nextflow can be loaded using modules.
$ module load nextflow
main.nf
Create a file, commonly called main.nf, containing the following content:
#!/usr/bin/env nextflow
wordcount="wordcount.py"
params.query="books/isles.txt"
println "I will count words of $params.query using $wordcount"
This main.nf file contains the main nextflow script that calls the processes (using channels and operators as
required). It does not need to be called main.nf but is standard practice.
Looking line by line:
#!/usr/bin/env nextflow
This is required to tell Nextflow it is a nextflow script.
wordcount="wordcount.py"
This sets a variable to refer to later.
params.query="books/isles.txt"
This is the query we have to find the number of words. Can be set with the commandline argument when running nextflow
with --query.
println "I will count words of $params.query using $wordcount"
This simply prints to screen using the variable and parameter above.
Running the code with:
$ nextflow run main.nf
Should produce:
N E X T F L O W ~ version 20.10.0
Launching `main.nf` [sad_mandelbrot] - revision: 43c1e227e3
I will count words of books/isles.txt using wordcount.py
A default work folder will also appear but will be empty.
Nextflow arguments
Try changing the variables and parameters on the commandline?
Solution
The parameter can changed with:
$ nextflow run main.nf --query "books/abyss.txt"Note the variable cannot be changed on the commandline.
nextflow.config
The nextflow.config is a file to set default global parameters for many parts of the nextflow components such as
parameters, process, manifest, executor, profiles, docker, singularity, timeline, report.
Create a nextflow.config. For now we will just use the config file to store the parameters.
params.query = "books/isles.txt"
params.wordcount = "wordcount.py"
params.outfile = "isles.dat"
params.outdir = "$PWD/out_dir"
Update the main.nf to remove the parameters and variables and use the values from nextflow.config in the println. You should end
up with:
#!/usr/bin/env nextflow
println "I will count words of $params.query using $params.wordcount and output to $params.outfile"
If this is run again it should output:
N E X T F L O W ~ version 20.10.0
Launching `main.nf` [angry_baekeland] - revision: ad46425e9c
I will count words of books/isles.txt using wordcount.py and output to isles.dat
Note we do not need to use the out_dir parameter yet.
params format
The
paramsin thenextflow.configcan be formatted as described or using:params { query = "books/isles.txt" wordcount = "wordcount.py" outfile = "isles.dat" }
Processes
Processes are what do work in Nextflow scripts. Processes require code to run, input data and output data (which may be used in another process).
In this example we want to run the wordcount.py code to code the number of words in an input file that creates a file
of word counts.
We will be using example data (the names of the files are used previously). The example data can be downloaded with:
$ curl -O https://raw.githubusercontent.com/ARCCA/intro_nextflow/gh-pages/files/nextflow-lesson.zip
$ unzip nextflow-lesson.zip
$ cd nextflow-lesson
Check the code works:
$ module load python
$ python3 wordcount.py books/isles.txt isles.dat
This should produce a file isles.dat with the word counts.
We should now be able to add this process to the main.nf. Copy the main.nf if not already there and add the
following:
process runWordcount {
script:
"""
module load python
python3 $PWD/wordcount.py $PWD/books/isles.txt isles.dat
"""
}
Note the use of $PWD to set the fullpath. This is crucial since Nextflow runs the code in another directory.
Run Nextflow again
$ nextflow run main.nf
And will produce:
N E X T F L O W ~ version 20.10.0
Launching `main.nf` [voluminous_coulomb] - revision: 6c143df13c
I will count words of books/isles.txt using wordcount.py and output to isles.dat
executor > local (1)
[38/2f5ad5] process > runWordcount [100%] 1 of 1 ✔
The isles.dat can be found in work directory such as:
$ tree work
work
└── 62
└── 9a9d076cc092ed99701282d3ee3e9f
└── isles.dat
The unique directories (using hashes) can be resumed by Nextflow.
Instead of hardcoding the arguments in the process, we can use the params and make sure $PWD is being used in
nextflow.config
Adding more parameters
Try adding more parameters to allow
python3to be called something else.Solution
Add to
nextflow.configthe parameterparams.appChangepythonto be$params.appparams.query = "$PWD/books/isles.txt" params.wordcount = "$PWD/wordcount.py" params.outfile = "isles.dat" params.outdir = "$PWD/out_dir" params.app = "python3"and the process is modified to be
$params.app $params.wordcount $params.query $params.outfile
Adding help functionality
Now that there is a series of arguments to the workflow it is beneficial to be able to print a help message.
In main.nf before the process definition the following can be added:
def helpMessage() {
log.info """
Usage:
The typical command for running the pipeline is as follows:
nextflow run main.nf --query \$PWD/books/isles.txt --outfile isles.dat
Mandatory arguments:
--query Query file count words
--outfile Output file name
--outdir Final output directory
Optional arguments:
--app Python executable
--wordcount Python code to execute
--help This usage statement.
"""
}
// Show help message
if (params.help) {
helpMessage()
exit 0
}
And add params.help to nextflow.config to a default value of false. Test the help functionality with:
$ nextflow run main.nf --help
We may want to add more dependencies that can be represented as:
You should now be able to run code and change what is run on the command line. We will now move onto learning about processes can use inputs and outputs.
Key Points
Nextflow describes your workflow in its Domain Specific Language (DSL)
Processes are executed independently, only way they can communicate is via asynchronous FIFO queues, called channels in Nextflow
Processes and channels
Overview
Teaching: 30 min
Exercises: 10 minQuestions
How do I pass information from processes?
Objectives
Understand processes.
Understand that channels link processes together.
Process more files
We have hopefully ran a simple workflow with fixed input and outputs than can be modified on the commandline. However to use the power of Nextflow, we should investigate further the concept of processes and how its link to the concept of channels.
Channels
Channels can act as a queue or a value.
- A queue is consumable, that is to say once it uses an entry in the channel it is removed.
- A value is fixed for the duration of the run, once a channel is loaded with a value it can be used over and over again by many processes.
When using channels the use of operators will be more apparent. operators link channels together or transform values in channels. For example we can add a channel to input many files.
Channel
.fromPath(params.query)
.set{queryFile_ch}
Notice the use of methods (...) and closures {...}
We can now add the channel as an input inside the process
input:
path(queryFile) from queryFile_ch
And change the process to include $queryFile instead of $params.query. This will provide no real advantage over the
current code but if we run with an escaped wildcard you can process as many files in the directory as required:
$ nextflow run main.nf --query $PWD/books/\*.txt
The output should now produce:
N E X T F L O W ~ version 20.10.0
Launching `main.nf` [sharp_euclid] - revision: 8594437a09
I will count words of /nfshome/store01/users/c.sistg1/nf_test/nextflow-lesson/books/*.txt using /nfshome/store01/users/c.sistg1/nf_test/nextflow-lesson/wordcount.py and output to isles.dat
executor > local (4)
[aa/dbc648] process > runWordcount (2) [100%] 4 of 4 ✔
Notice the 4 of 4. This has run the process 4 times for each of the 4 books we have available.
Output names
Can you see one issue though? The output file does not change. We need to provide a sensible output name. Try looking inside the
workdirectory. How does multiple jobs stored?Solution
Each job has its own unique directory. THerefore no files are overwritten even if each job has the same output name.
To give the output name something sensible we could use ${queryFile.baseName}.dat as the output. Try it and see if it
works and outputs the files in the current directory.
Bioinformatics specific functions
Nextflow was designed from a bioinformatics perspective so there are some useful functions.
For example splitting a fasta file is common practice so can be done with:
Channel .frompath(params.query) .splitFasta(by: 1, file:true) .set { queryFile_ch }This will split the fasta file into chunks of size 1 and create individual processes for each chunk. However you will need to join these back again. Nextflow comes to the rescue with:
output: path(params.outFileName) into blast_output_chAnd after the process use the channel
blast_output_ch .collectFile(name: 'combined.txt', storeDir: $PWD)
Adding more processes
Back to the book example. We can now process all the books but what can we do with the results. There is something call Zipf’s Law. We can use the output from each of the wordcounts from the book to test Zipf’s Law. For example the following dependency diagram shows what we would like to achieve:
Zipf’s Law
Zipf’s Law is an empirical law formulated using mathematical statistics that refers to the fact that many types of data studied in the physical and social sciences can be approximated with a Zipfian distribution, one of a family of related discrete power law probability distributions.
Zipf’s law was originally formulated in terms of quantitative linguistics, stating that given some corpus of natural language utterances, the frequency of any word is inversely proportional to its rank in the frequency table. For example, in the Brown Corpus of American English text, the word the is the most frequently occurring word, and by itself accounts for nearly 7% of all word occurrences (69,971 out of slightly over 1 million). True to Zipf’s Law, the second-place word of accounts for slightly over 3.5% of words (36,411 occurrences), followed by and (28,852). Only 135 vocabulary items are needed to account for half the Brown Corpus.
Source: Wikipedia:
Let’s create a new process that runs the Python code to test Zipf’s Law. You cansee what it produces by running it on the command line.
$ python zipf_test.py abyss.dat isles.dat last.dat > results.txt
This can be added as a process with:
process testZipf {
script:
"""
module load python
python3 zipf_test.py abyss.dat isles.dat last.dat > results.txt
"""
}
However we have to define the inputs.
In the original process the output was just a name, however we can now add the ouput to a channel. We now do not need
to store the .dat output in $PWD. We can leave them in the work directories. (Remember to change the original
RunWordcount process).
output:
path("${queryFile.baseName}.dat") into wordcount_output_ch
The input for testZipf will use the collect() method of the channel:
input:
val x from wordcount_output_ch.collect()
script:
"""
module load python
python3 zipf_test.py ${x.join(" ")} > $PWD/results.txt
"""
Lets run it and see what happens.
$ nextflow run main.nf --query $PWD/books/\*.txt
N E X T F L O W ~ version 20.10.0
Launching `main.nf` [lethal_cuvier] - revision: 4168a6dfe0
I will count words of /nfshome/store01/users/c.sistg1/nf_test/nextflow-lesson/books/*.txt using /nfshome/store01/users/c.sistg1/nf_test/nextflow-lesson/wordcount.py and output to isles.dat
executor > local (5)
[0c/46bef2] process > runWordcount (2) [100%] 4 of 4 ✔
[13/001d31] process > testZipf [100%] 1 of 1 ✔
This now runs two processes. Output should be stored in results.txt within the current directory. We should really
store the results in the outdir location defined in params. The publishDir directive defines a location to make
available
output from a process.
publishDir "$params.outdir"
By defining the output for the process it will know to copy the data there.
Configure the output directory
Currently we are just outputting to
$PWD. How can this be modified. Try using thepublishDirdirective in thetestZipfprocess and define an output file to copy to it.Solution
We first set
publishDirto the value inparams.publishDir "$params.outdir"Then define an output that will be available in
publishDir.output: path('results.txt')
We have now created a very simple pipeline where four processes run in parallel and a process uses all output to create
a results file. We shall now move to looking closer at the nextflow.config.
Key Points
Channels and processes are linked, one cannot exist without the other.
Advanced configuration
Overview
Teaching: 15 min
Exercises: 5 minQuestions
What functionality and modularity to Nextflow?
Objectives
Understand how to provide a timeline report.
Understand how to obtain a detailed report.
Understand configuration of the executor.
Understand how to use Slurm
We have got to a point where we hopefully have a working pipeline. There is now some configuration options to allow us to explore and modify the behaviour of the pipeline.
The nextflow.config was used earlier to set parameters for the Nextflow script. We can also use it to set a number of
different options.
Timeline
To obtain a detailed timeline report add the following to the nextflow.config
timeline {
enabled = true
file = "$params.outdir/timeline.html"
}
Notice the user of $params.outdir that can be defined in the params section to a default value such as
$PWD/out_dir.
The timeline will look something like:
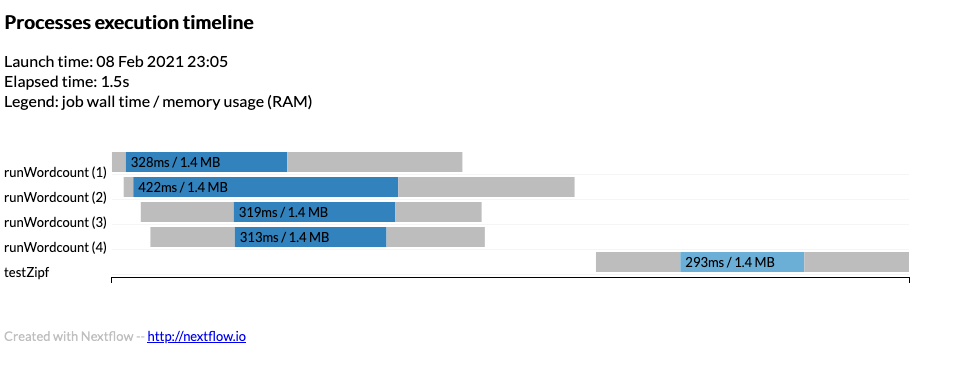
Example timeline can be found timeline.html
Report
A detailed execution report can be created using:
report {
enabled = true
file = "$params.outdir/report.html"
}
Example can be found report.html
Executors
If we are using a job scheduler where user limits are in place we can define thise to stop Nextflow abusing the
scheduler. For example to report the queueSize as 100 and submit 1 job every 10 seconds we would define the executor
block as:
executor {
queueSize = 100
submitRateLimit = '10 sec'
}
Profiles
To use the executor block as described previously then a profile can be used to define a job scheduler. Within the
nextflow.config file define:
profiles {
slurm { includeConfig './configs/slurm.config' }
}
and within the ./configs/slurm.config define the Slurm settings to use:
process {
executor = 'slurm'
clusterOptions = '-A scwXXXX'
}
Where scwXXXX is the project code to use.
This can be used on the command line:
$ nextflow run main.nf -profile slurm
Or the whole definition can be defined within the process we can define the executor and cpus
executor='slurm'
cpus=2
You can also define the profile on the command line but add to existing profile such as using -profile slurm when
running but setting cpus = 2 in the process. Note for MPI codes you would need to put clusterOptions = '-n 16' for
a 16 tasks to use for MPI. Be careful not to override options such as clusterOptions that define the project code.
Manifest
A manifest can describe the workflow and provide a Github location. For example
manifest {
name = 'ARCCA/intro_nextflow_example'
author = 'Thomas Green'
homePage = 'www.cardiff.ac.uk/arcca'
description = 'Nextflow tutorial'
mainScript = 'main.nf'
version = '1.0.0'
}
Where the name is the location on Github and mainScript is the location of the file (default in main.nf).
Try:
$ nextflow run ARCCA/intro_nextflow_example --help
To update from remote locations you can run:
$ nextflow pull ARCCA/intro_nextflow_example
To see existing remote locations downloaded:
$ nextflow list
Finally, to print information about remote you can:
$ nextflow info ARCCA/intro_nextflow_example
Labels
Labels allow to select what the process can use from the nextflow.config or in our case the options in the Slurm
profile in ./configs/slurm.config
process {
executor = 'slurm'
clusterOptions = '-A scw1001'
withLabel: python { module = 'python' }
}
Defining a process with the above label 'python' and will load the python module.
Modules
Modules can also be defined in the process (rather than written in the script) with the module directive.
process doSomething {
module = 'python'
"""
python3 --version
"""
}
Conda
Conda is a userful software installation and management system and commonly used by various topics. There are a number of ways to use it.
Specify the packages.
process doSomething {
conda 'bwa samtools multiqc'
'''
bwa ...
'''
Specify an environment.
process doSomething {
conda '/some/path/my-env.yaml'
'''
command ...
'''
Specify a pre-existing installed environment.
process doSomething {
conda '/some/path/conda/environment'
'''
command ...
'''
It is recommended to use conda inside a profile due to there might be another way to access the software such as via docker or singularity.
profiles {
conda {
process.conda = 'samtools'
}
docker {
process.container = 'biocontainers/samtools'
docker.enabled = true
}
}
Generate DAG
From the command line a Directed acyclic graph (DAG) can show the dependencies in a nice way. Run nextflow with:
$ nextflow run main.cf -with-dag flowchart.png
The flowchart.png will be created and can be viewed.
Hopefully the following page has helped you understand the options to dig deeper into your pipeline and maybe make it more portable by using labels to select what to do on a platform. Lets move onto running Nextflow on Hawk.
Key Points
Much functionality is available but had to be turned on to use it.
Running on cluster
Overview
Teaching: 15 min
Exercises: 15 minQuestions
What configuration should I use to run on a cluster?
Objectives
Understand how to use Slurm specific options
Make sure the correct filesystem is used.
We should now be comfortable running on a single local machine such as the login node. To use the real power of a supercomputer cluster we should make sure each process is given the required resources.
Executor
Firstly the executor scope provides control how things are run. For example for slurm we can set a limit to the number of jobs in the queue
executor {
name = 'slurm'
queueSize = 20
pollInterval = '30 sec'
}
Profile
In the previous section we encountered the concept of a profile. Lets look at that again but call the profile cluster and include executor and process.
Lets ignore the includeConfig command for now.
profiles {
cluster {
executor {
name = 'slurm'
queueSize = 20
pollInterval = '30 sec'
}
process {
executor = 'slurm'
clusterOptions = '-A scw1001'
}
}
}
This defines a profile to set options in a executor and process, this submits to Slurm using sbatch and passes the clusterOptions, in this case the
project code used to track the work.
Specifying resource
The default profiles method in Nextflow handles many things out of the box, for example the slurm executor has the
following available:
- cpus - the number of cpus to use for the process, e.g.
cpus = 2 - queue - the partition to use in Slurm, e.g.
queue = 'htc' - time - the time limit for the job, e.g.
time = 2d1h30m10s) - memory - the amount of memory used for process, e.g.
memory = '2 GB' - clusterOptions - the specific arguments to pass to
sbatch, e.g.-A scwXXXX
As you can see most of the usual subjects are there, however if MPI jobs were ever run inside Nextflow the
clusterOptions would need to be used to define the number of MPI tasks with -n and the --ntasks-per-node. Same
for if GPUs were to be used with --gres=gpu
To submit to Slurm we can run the following:
$ nextflow run main.nf -profile cluster
Submission to Slurm
With your current pipeline you should have 4 processes that can run in parallel and a process that depends on all 4 processes to finish. Create a Slurm profile and submit your pipeline. Watch the jobs queue in Slurm with
squeue -u $USER.Solution
Following the advice you should be able to use many of the defaults since we are not using parallel code so
ncpus = 1will be the default. Just make sureclusterOptions = "-A scwXXXX"to specify your project code.
Work directory
The work directory where processes are run is by default in the location where you run your job. This can be changed by
environment variable (not very portable) or by configuration variable. In nextflow.config set the following:
workDir = "/scratch/$USER/nextflow/work"
This will set a location on /scratch to perform the actual work.
The previous option publishDir by default symlinks the required output to a convenient location. This can be changed
by specifying the copy mode.
publishDir "$params.outdir", mode: 'copy'
Filesystems
Modify the pipeline in
main.nfto use a working directory in your/scratchspace and make the data available in yourpublishDirlocation with a new mode such as copy.Solution
The solution should be straight forward using information above.
In
nextflow.config:workDir = "/scratch/$USER/nextflow/work"In
main.nf:publishDir "$params.outdir", mode: 'copy'
Module system
The module system on clusters is a convenient way to load common software. As described before the module can be loaded
by Nextflow before the script is run using the module directive.
module 'python'
This will load the python module which can be loaded normally with
$ module load python
To load multiple modules such as python and singularity separate with a colon
module 'python:singularity'
This has hopefully given you some pointers to configure your Nextflow environment for the cluster. One aspect of the cluster is the use of Singularity to simplify sharing of specific builds of code. Nextflow support the tool Singularity to manage this feature which will shall look at in the next section.
Key Points
Tweak your configuration to make the most of the cluster.
Singularity
Overview
Teaching: 15 min
Exercises: 15 minQuestions
How can I quickly reproduce or share my pipelines?
Objectives
Understand how Nextflow uses Singularity to perform tasks.
Use Singularity to perform a simple pipeline process.
Having setup a job using locally installed software, we can instead use software distributed in containers. Containers allow for easier management of software due to being able to run the same code across many systems, usually code is required to be rebuilt for each operating system and hardware.
The most common ways to run containers is to either use Docker and Singularity. Docker can suffer from some issues running on a supercomputer cluster, so it may have Singularity installed to allow containers to run. We will be using Singularity.
Singularity training
ARCCA provides a Singularity course if more detailed information how to use Singularity is required. See list of training courses.
Using Singularity
Singularity can be specified in the profiles section of your configuration such as:
profiles {
slurm { includeConfig './configs/slurm.config' }
singularity {
singularity.enabled = true
singularity.autoMounts = true
}
}
To specify the container to use we can specify it within the process in our pipeline.
process runPython {
container = 'docker://python'
output:
stdout output
script:
"""
python --version 2>&1
"""
}
output.subscribe { print "\n$it\n" }
Note the use of subscribe which allows a function to be used to everytime a value is emitted by the source channel.
The above should output:
$ nextflow run main.nf -profile singularity
N E X T F L O W ~ version 20.10.0
Launching `main.nf` [spontaneous_spence] - revision: 8c50b57f4e
executor > local (1)
[59/cd3d1e] process > runPython [ 0%] 0 of 1
Python 3.9.1
executor > local (1)
[59/cd3d1e] process > runPython [100%] 1 of 1 ✔
Notice the change in Python version.
Compare runs.
Using the simple singularity pipeline, investigate what happens when the
containerdirective is removed or commented out.Solution
You should see a change in version number when commenting out the container.
Python 2.7.5
Using Singularity can remove some of the head-aches of making sure the correct software is installed. If an existing pipeline has a container then it should be possible to run it within Singularity inside Nextflow. Creating your own container should also be possible if required to share with others.
Having had a bit of a detour to explore some extra options we now go back to the original workflow and explore a few final things with it.
Key Points
Singularity allows for quick reproduction of common tasks that others have published.
Wrapping up and where to go
Overview
Teaching: 15 min
Exercises: 15 minQuestions
Where do I go now?
Objectives
Understand the basics in Nextflow pipelines and the options available.
Understand the next steps required.
The pipeline
Using the -with-dag option we can see the final flowchart
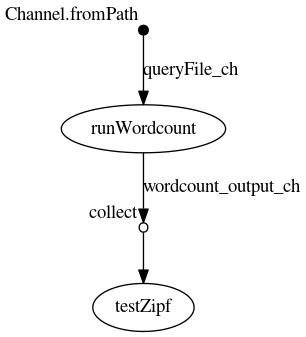
This doesnt show the particular runtime options but the raw dependency of the given processes in the Nextflow script.
This section shall document some of the questions raised in sessions.
Issues raised
- Chunking up data from channels.
When a list of files is send into a channel, a process can be defined to read an entry and process that one file. A case was raised where we may want to send 300 files to the channel and for a process to read 10 files from the channel at a time and perform a average on the data in the files. This can be achieved with the:
Channel
.from( 1,2,3,4,5,6 )
.take( 3 )
.subscribe onNext: { println it }, onComplete: { println 'Done' }
This example is taken from the documentation for Nextflow. This will produce:
1
2
3
Done
We could easily use this within our pipeline to process 2 files in the same process for example:
input:
tuple path(queryFile1), path(queryFile2) from queryFile_ch.collate(2)
output:
tuple path("${queryFile1.baseName}.dat"), path("${queryFile2.baseName}.dat") into wordcount_output_ch
script:
"""
python3 $PWD/../wordcount.py $queryFile1 ${queryFile1.baseName}.dat
python3 $PWD/../wordcount.py $queryFile2 ${queryFile2.baseName}.dat
"""
This will emit a tuple (people familiar with Python should know this term, think of it as a fixed list). Note the use
of collate on the input to the process. The output uses tuple to send both files to the output. A change is
required in the receiving process testZipf
input:
val x from wordcount_output_ch.flatten().collect()
The output from the run will look like:
$ nextflow run main.nf --query $PWD/../books/\*.txt
N E X T F L O W ~ version 20.10.0
Launching `main.nf` [magical_bardeen] - revision: dae9352cb9
I will count words of /nfshome/store01/users/c.sistg1/nf_training_Feb21/nextflow-lesson/work/../books/*.txt using wordcount.py and output o isles.dat
executor > local (3)
[54/2d1cdb] process > runWordcount (1) [100%] 2 of 2 ✔
[8b/217add] process > testZipf [100%] 1 of 1 ✔
This will first flatten the tuples into a list and then collect will emit all entries as a list to the val x.
Hopefully this shows the power of Nextflow is dealing with specific use cases.
- Self-learning
Due to time constraints during the sessions, opportunities to explore Nextflow at your own pace is limited. It would be good to setup a Nextflow community within the University to promote the use and encourage working together in solving common issues.
Interfaces to Nextflow
There are currently two interfaces to Nextflow that are being looked at:
- Nextflow Tower
Nextflow Tower is promoted by the developers of Nextflow. The cloud version is potentially a risk to store access keys to local resource such as the University supercomputer. There is a community edition on Github that suggests single users can run. Further investigation is required.
- DolphinNext
DolphinNext is an open project to provide an interface to Nextflow similar to Nextflow Tower. ARCCA currently has a test system up which we will go through during the live sessions. If users are interested in this please get in contact with us.
A tutorial to use DolphinNext is available on Github
Links to other resources
Key Points
Ask for help and support if you require assistance in writing pipelines on the supercomputer. The more we know people using it the better.